Plant Biotech, Climate Change and Sustainability
Our collective research endeavors focus on understanding and mitigating the impacts of environmental change across diverse biological systems. From the intricate molecular mechanisms that enable plants to withstand drought and salinity, to the neural circuits guiding insect pollinators, the microbial communities degrading pollutants, and the cellular pathways maintaining protein homeostasis under extreme conditions, our work explores the fundamental principles of adaptation and resilience. We investigate the genetic and ecological foundations of biodiversity, examining how organisms from aquatic ecosystems to wetlands and terrestrial environments respond to stressors like climate change, pollution, and habitat loss.
Employing a wide range of cutting-edge techniques, including high-throughput sequencing, advanced bioinformatics, statistical modeling and game theory, metagenomics and synthetic biology, conservation genetics, neuroethology, microbial genomics, and plant biotechnology, we seek to unravel the complex interactions between organisms and their environments. We aim to translate our fundamental discoveries into practical solutions for sustainable agriculture, conservation, and bioremediation. By understanding the molecular basis of plant stress tolerance, we strive to enhance crop resilience and ensure food security. By studying the genetics and ecology of aquatic species, we contribute to the conservation of vital ecosystems. By elucidating the neurobiology of insect pollinators, we safeguard essential ecological services. And by harnessing the power of microbial communities, we develop innovative strategies for environmental cleanup.
Ultimately, our research is driven by a shared commitment to understanding the biological responses to global change and developing strategies that promote ecological sustainability and human well-being. We recognize that the challenges facing our planet are interconnected and that a holistic, interdisciplinary approach is essential for finding lasting solutions.

Dr. Neelesh Dahanukar
Assistant Professor
Department of Life Sciences
[email protected]
Research focus
- Conservation genetics and genomics for understanding the molecular ecology of threatened species and their conservation needs.
- Population dynamics and exploitation analysis for conservation management and action.
- Evolutionary game theory and conflict resolution among various stakeholders.

Dr. Koyeli Mapa
Associate Professor
Department of Life Sciences
[email protected]
Research focus
- Proteotoxic stresses due to several altered environmental conditions like extreme heat stress, chemical stresses, or DNA-damaging stresses.
- Modulation of cellular response by endoplasmic reticulum-unfolded protein response pathway.

Dr. Richa Priyadarshini
Associate Professor
Department of Life Sciences
[email protected]
Research focus
- Whole genome sequencing of Novel Bacteria.
- Harnessing biotechnological applications of novel bacterial isolates.
- Studying the role of microbes in plastic degradation.

Dr. Rohini Garg
Associate Professor
Department of Life Sciences
[email protected]
Research focus
- Epigenomic regulation of abiotic stress response in chickpea.
- Gene regulation under abiotic stress response in chickpea.
- Small peptide-mediated stress tolerance in chickpeas and Arabidopsis.
- Genome and epigenome editing of regulatory regions in Arabidopsis and chickpea.

Dr. Tanvi Deora
Faculty Fellow
Department of Life Sciences
[email protected]
Research focus
- Sensory Ecology: We study the insect nervous system to understand how sensing and learning affect plant pollination.
- Climate change and Anthropogenic Impact on Pollination: We study the pollination networks in the field to ask how various environmental factors such as light pollution, and decreasing flower diversity impact the pollination networks of diurnal and nocturnal insects.
Stress Survival: The Plant's Molecular Toolkit
Dr. Rohini Garg
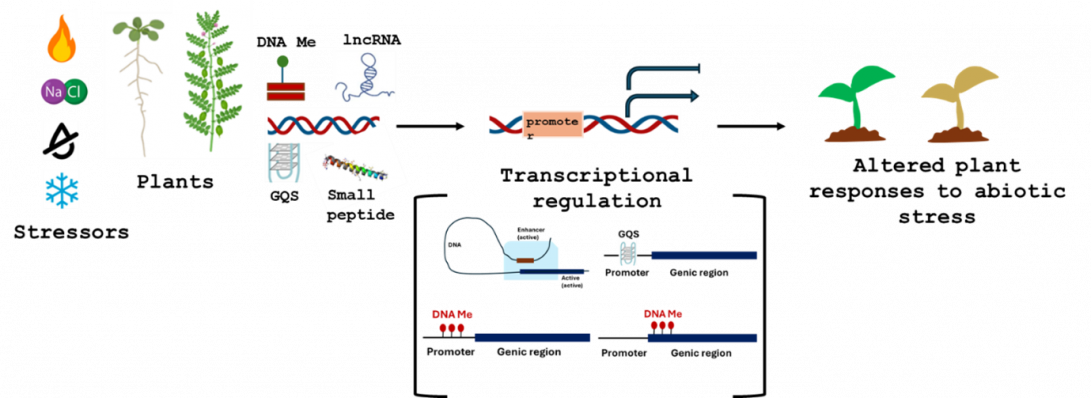
Plants are subjected to a variety of abiotic stresses like cold, heat, drought, and salinity. We study molecular responses to abiotic stresses in Arabidopsis and chickpea, focusing on DNA methylation, lncRNAs, DNA secondary structures, and small peptides. Our goal is to identify target genes for enhancing plant stress tolerance.
Plants face constant threats from environmental stresses like drought and salinity. We study the molecular regulation of abiotic stress tolerance in plants, focusing on chickpea and Arabidopsis responses to drought and salinity. Our research delves into the roles of DNA methylation, DNA secondary structures, long non-coding RNAs, and small peptides in modulating gene expression under stress. We utilize high-throughput sequencing (e.g., RNA-seq, ChIP-seq), advanced bioinformatics analyses, and biotechnological approaches like gene editing and overexpression to decipher these complex regulatory networks. By integrating cutting-edge sequencing technologies with sophisticated bioinformatics and targeted genetic manipulation, we aim to understand the fundamental principles of plant adaptation and translate this knowledge into improved crop varieties. Ultimately, we seek to identify and manipulate specific genes to enhance crop resilience in challenging environments.
Lab homepage: https://snu.edu.in/faculty/rohini-garg/
Relevant publications:
- Ali N, Singh S and Garg R (2025). Unlocking Crops' Genetic Potential: Advances in Genome and Epigenome Editing of Regulatory Regions. Current opinion in Plant Biology (accepted). 83, 102669 https://doi.org/10.1016/j.pbi.2024.102669. (IF = 8.3)
- Garg R., Sahu SK, Jain M, (2024) Single same-cell multiome for dissecting key plant traits. Trends in Plant Sciences, online DOI: 10.1016/j.tplants.2024.10.008. Spotlight article (IF = 17.3)
- Singh V, Gupta K, Singh S, Jain M, Garg R. (2023) Unravelling the molecular mechanism underlying drought stress response in chickpea via integrated multi-omics analysis. Frontiers in Plant Sciences, 14. (*corresponding author). (IF = 5.6)
- K Gupta, R Garg. (2023). Unravelling Differential DNA Methylation Patterns in Genotype Dependent Manner under Salinity Stress Response in Chickpea. IJMS, 24 (3), 1863. (*corresponding author). (IF = 5.6)
- R Garg, PK Subudhi, RK Varshney, M Jain (2023). Abiotic stress: Molecular genetics and genomics, volume II. FiPS, Abiotic stress: molecular genetics and genomics, Volume II 16648714, 7. (IF = 5.6)
- Jain M, Bansal J, Rajkumar MS, Garg R (2022). An integrated transcriptome mapping the regulatory network of coding and long non-coding RNAs provides a genomics resource in chickpea. Commun Biol. 19;5(1):1106. (IF = 5.9)
- Jain M, Garg R (2021). Enhancers as potential targets for engineering salinity stress tolerance in crop plants. Physiologia Plantarum. 173(4):1382-1391. (IF = 6.4)
Sustainability and Conservation of Life Below Water
Dr Neelesh Dahanukar
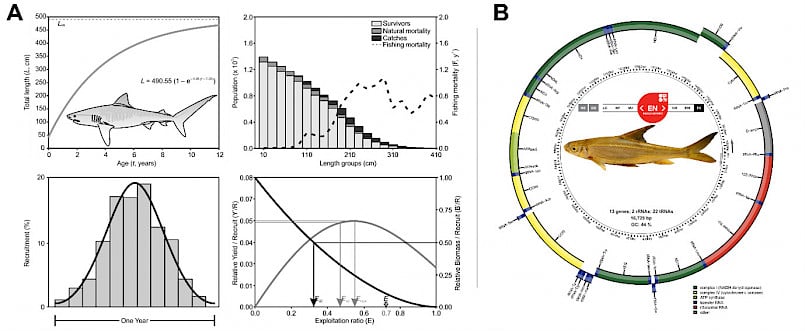
Conservation ecology of aquatic fauna. (A) Population dynamics and exploitation of Tiger Sharks shows its fisheries in Arabian sea is not sustainable and needs immediate conservation action. (B) Mitogenome of Endangered endemic fish Lepidopygopsis typus revealed that some of its genes are under relaxed selection. Restricted distribution, low population size and relaxation in selection can likely trigger extinction in L. typus, and therefore urgent conservation and monitoring plans are required.
Sustainable Development Goal (SDG) 14, Life Below Water, calls for conservation and sustainable use of the world’s oceans, seas, and marine resources and can also be extended to include freshwater ecosystems. We employ a variety of research methods including conservation genetics, high throughput genomics analysis, statistical inferences, and mathematical modelling to understand the conservation needs for marine and freshwater fishes (primary interest) and other vertebrate and invertebrate taxa. Work on conservation genetic and genomics include population genetics analysis for understanding genetic heterogeneity, dispersal and fragmentation, historical population changes, genome wide selection patterns and molecular systematics to understand conservation needs for various threatened taxa. We also employ statistical methods to understand the sustainability of exploited fisheries of threatened species, both marine and freshwaters, to understand the future of natural stocks and propose conservation action and management plans. We use game theoretical analysis to understand how to resolve conflict among various stakeholders for ensuring conservation of natural resources.
Laboratory homepage: https://sites.google.com/snu.edu.in/molecoevo
Relevant publications:
- Gurugubelli, M., Abisha, C., Arundhathy, T.A., Ranjeet, K., Dahanukar, N.*, & Raghavana, R.* (Accepted) A megafauna in distress: characterizing an unsustainable fishery for tiger sharks in the Arabian Sea. Biological Conservation.
- Chandra, S., Abhilash, R., Sidharthan, A., Raghavan, R. & Dahanukar, N.* (2024) Complete mitogenome of Lepidopygopsis typus, an evolutionarily-distinct, endangered cyprinid fish from the Western Ghats Biodiversity Hotspot: phylogenetic relationships and implications for conservation. Gene 898: 148098. https://doi.org/10.1016/j.gene.2023.148098
- Raghavan, R., Britz, R. & Dahanukar, N. (2021) Poor groundwater governance threatens ancient subterranean fishes. Trends in Ecology & Evolution 36(10): 875-878. https://doi.org/10.1016/j.tree.2021.06.007
- Joshi, P., Dahanukar, N., Bharade, S., Dethe, V., Dethe, S., Bhandare, N. & Watve, M. (2021) Combining payment for crop damages and reward for productivity to address wildlife conflict. Conservation Biology 35(6): 1923-1931. https://doi.org/10.1111/cobi.13746
Neuromechanics and ecology of insect pollination behaviors
Dr Tanvi Deora
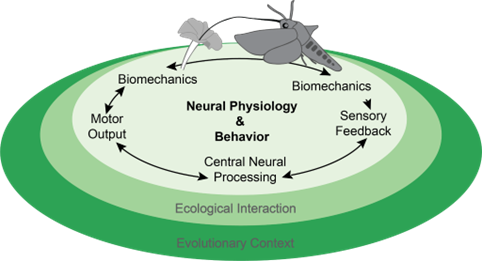
The Deora lab studies the neural and mechanical basis of insect pollination in lab and in the field to understand how neural dynamics affect animal behavior and pollination service. We also study how the natural history and ecology of an animal shape’s its neural system and behavior.
From the tiny jellyfish in the sea to large birds in the sky, nervous systems of animals shape their interactions with their environment. In our lab, we study how insects' nervous systems help them interact with flowers during pollination. Insects are the major pollinators of flowering plants and how they find and feed from flowers have a huge impact on their role as pollinators. Several sensory cues such as vision, olfaction, hygrosensation (humidity), touch etc. shape these crucial interactions. In the lab we study how sensory systems of insects like butterflies and moths guide locomotion to enable tasks that demand expert control such as targeting a tiny nectary opening in the flower to drink nectar while also hovering and hence pollinating the flowers. We study the impact of sensing and learning on feeding and pollination efficiency to reveal sensorimotor mechanisms underlying pollination interactions. Combining comparative studies in the lab and field, we hope to understand how natural history of animals have shaped their nervous systems and their ecological interactions. Importantly, these studies will help us understand how declining floral and insect biodiversity or increasing anthropogenic pollutants such as artificial light at night impact ecological services like insect pollination.
Laboratory homepage: https://sites.google.com/view/tanvideora/home
Relevant publications:
- Deora T, Ahmed MA, Daniel TL, Brunton BW. Tactile active sensing in an insect plant pollinator. Journal of Experimental Biology. 2021 Feb 15;224(4):jeb239442.
- Deora T, Ahmed MA, Brunton BW, Daniel TL. Learning to feed in the dark: how light level influences feeding in the hawkmoth Manduca sexta. Biology Letters. 2021 Sep 15;17(9):20210320.
- Stöckl A, Deora T. The hawkmoth proboscis: an insect model for sensorimotor control of reaching and exploration. Integrative and Comparative Biology. 2024 Nov;64(5):1354-70.
- Stanchak KE, Deora T, Weber AI, Hickner MK, Moalin A, Abdalla L, Daniel TL, Brunton BW. Intraspecific Variation in the Placement of Campaniform Sensilla on the Wings of the Hawkmoth Manduca Sexta. Integrative Organismal Biology. 2024;6(1):obae007.
- Aiello BR, Stanchak KE, Weber AI, Deora T, Sponberg S, Brunton BW. Spatial distribution of campaniform sensilla mechanosensors on wings: form, function, and phylogeny. Current Opinion in Insect Science. 2021 Dec 1;48:8-17.
Harnessing Microbial Biodiversity for biodegradation of plastics
Dr Richa Priyadarshini
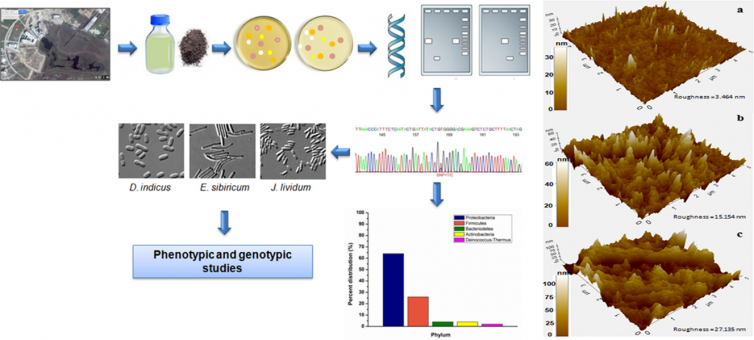
Chauhan D, Agrawal G, Deshmukh S, Roy SS, Priyadarshini R. Biofilm formation by Exiguobacterium sp. DR11 and DR14 alter polystyrene surface properties and initiate biodegradation. RSC Adv. 2018 Nov 8;8(66):37590-37599. doi: 10.1039/c8ra06448b.
Wetlands, defined as land areas that are saturated with water – either permanently or seasonally – are rich ecosystems supporting a wide array of flora and fauna. The rich biodiversity of wetlands has attracted attention due to the high number of species present in the small geographical area. Bacterial diversity in wetland ecosystems is estimated to be an amalgamation between aquatic and terrestrial habitats. Metagenomics and synthetic biology advancements have paved the way for utilizing microbial diversity for tailored applications. Harnessing microbial biodiversity supports sustainable development by reducing environmental impact and promoting circular bioeconomy principles. One of the primary goals of our research is to isolate and characterize microbes with biotechnological potential from wetlands.
Biodegradation is a process by which microbial organisms that transform or degrade chemicals introduced into the environment. Our laboratory has isolated two bacterial strains from Dadri wetlands, Exiguobacterium sibiricum strain DR11, Exiguobacterium undae strain DR14, with potential degrade plastic, especially polystyrene. These bacterial strains establish biofilms on plastic material and secrete enzymes probably initiating the process of depolymerisation. Growth studies suggested that these Exiguobacterium strains can utilize polystyrene as a carbon source. Further characterization of these bacteria would provide insight into the enzymes and pathways used in plastic biodegradation.
Lab homepage: https://snu.edu.in/faculty/richa-priyadarshini/
Relevant Publications:
- Shrivaishnavi Ranganathan, Deepa Sethi, Sandhya Kasivisweswaran, L Ramya , Richa Priyadarshini*, Ragothaman M Yennamalli*. Structural and functional mapping of ars gene cluster in Deinococcus indicus 2022. Comput Struct Biotechnol J. 21:519-534. doi: 10.1016/j.csbj.2022.12.015
- Richa Priyadarshini, Thava Palanisami, Arulazhagan Pugazhendi, Arumugam Gnanamani Obulisamy Parthiba Karthikeyan. Editorial: Plastic to Bioplastic (P2BP): A Green Technology for Circular Bioeconomy. 2022. Front Microbiol. 2022 Apr 18;13:851045. doi: 10.3389/fmicb.2022.851045. eCollection 2022.
- Deepa Sethi, Richa Priyadarshini*. Isolation, Propagation, and Identification of Bacterial Species with Hydrocarbon Metabolizing Properties from Aquatic Habitats. 2021. J Vis Exp. Dec 7;(178). doi: 10.3791/63101.
- Deepika Chauhan, Pulkit Anupam Srivastava, Barbara Ritzl, Ragothaman M Yennamalli, Felipe Cava and Richa Priyadarshini*. Amino Acid-Dependent Alterations in Cell Wall and Cell Morphology of Deinococcus indicus DR1. July 2019. Frontiers in Microbiology. 10:1449.
- Chauhan, Deepika; Agrawal, Guncha; Deshmukh, Sujit; Roy, Susanta and Richa Priyadarshini, 2018. Biofilm formation by Exiguobacterium DR11 and DR14 alter polystyrene surface properties and initiate biodegradation. RSC Adv., 8, 37590
- Deepika Chauhan, Pulkit Srivastava, Ragothaman Yennamalli, and Richa Priyadarshini. 2017 Draft Genome Sequence of Deinococcus indicus DR1, a Novel Strain Isolated from a Freshwater Wetland. Genome Announc. Aug 3;5 (31).
Understanding organelle-specific protein homeostasis during environmental stresses
Dr Koyeli Mapa
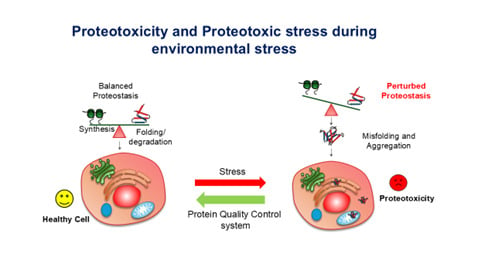
Our laboratory is working towards understanding how organisms cope with protein misfolding stresses, also known as proteotoxic stresses, due to several altered environmental conditions like extreme heat stress, chemical stresses, or DNA-damaging stresses. In the rapidly changing world with growing concerns about global warming and environmental pollution, organisms are constantly facing the challenges of maintaining the functional proteome within cells. We are experimentally simulating such stresses in laboratory conditions and trying to understand the response and survival pathways adopted by different organisms to endure such stresses.
Lab homepage: https://koyelimapa.wixsite.com/protein-homeostasis
Relevant Publications:
- Sse1, Hsp110 chaperone of yeast, controls the cellular fate during endoplasmic reticulum stress. Jha MP, Kumar V, Ghosh A, Mapa K* (*corresponding authors). G3, Genes|Genomes|Genetics (Bethesda). 2024 Jun 5;14(6):jkae075. doi: 10.1093/g3journal/jkae075.
- Stress responses elicited by misfolded proteins targeted to mitochondria. Rao KBN, Pandey P, Sarkar R, Ghosh A, Mansuri S, Ali M, Majumder P, Kumar KR, Ray A, Raychaudhuri S and Mapa K* (*corresponding author). J Mol Biol. 2022 Jun 30;434(12):167618. doi: 10.1016/j.jmb.2022.167618.
- Conserved and divergent chaperoning effects of Hsp60/10 chaperonins on protein folding landscapes. Sadat A, Tiwari S, S Sunidhi, Chaphalkar A, Kocharr M, Ali M, Zaidi Z, Sharma A, Verma K, Rao KBN, Tripathi M, Ghosh A, Gautam D, Atul, Ray A*, Mapa K*, Chakraborty K* (*corresponding authors). Proc Natl Acad Sci U S A. 2022 May 3;119(18):e2118465119. doi: 10.1073/pnas.2118465119.
Endoplasmic reticulum-unfolded protein response pathway modulates the cellular response to mitochondrial proteotoxic stress. Sarkar R, Rao KBN, Jha MP, Mapa K* (*corresponding author). Cell Stress Chaperones. 2022 May;27(3):241-256. doi: 10.1007/s12192-022-01264-2.