Disease Pathogenesis and Therapeutics Group
The Disease Pathogenesis and Therapeutics Group at the Department of Life Sciences is committed to understanding the causes and effects of both communicable and non-communicable diseases. Their goal is to identify effective treatment methods that enhance clinical outcomes. This research aligns with the United Nations' Sustainable Development Goal (SDG) 3, which aims to ensure health and well-being for all.
Multiple government and industrial funding sources, including the Department of Biotechnology, Department of Science and Technology, Council of Research and Engineering Board, Indian Council of Medical Research, and L’Oréal, generously support the research programs.
The members of this group are:
Dr. Sanjeev A Galande, Dean, School of Natural Sciences
Dr. Colin C Jamora, Senior Professor and Head of the Department
Dr. Sri Krishna Jayadev M, Associate Professor
Dr. Richa Priyadarshini, Associate Professor
Dr. Naga Suresh Veerpau, Associate Professor
Dr. Asish Gupta, Associate Professor
Dr. Anindita Chakrabarty, Associate Professor
Dr. Rajan Vyas, Assistant Professor, Life Sciences
The department's cutting-edge infrastructure, multiple core facilities and the renowned Center of Excellence in Epigenetics are crucial in enhancing the group's impactful research activities. This exceptional support empowers us to push the boundaries of science and make significant advancements in the field.

https://doi.org/10.1021/acs.jnatprod.2c00521
Sanjeev Galande:
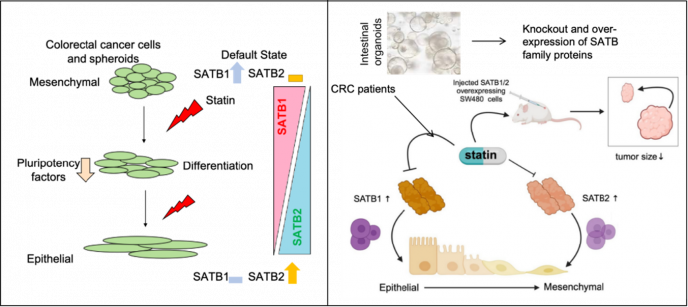
Summary: Colorectal cancer (CRC) is the third leading cause of cancer-related deaths worldwide. The chromatin landscape is crucial in gene regulation, particularly in complex diseases like cancer. SATB1, a key chromatin organizer, regulates gene expression and has been identified as a novel target of Wnt/β-catenin signaling in CRC. Our lab investigates the role of SATB1/SATB2 in CRC progression, metastasis, and tumor-initiating cells (TICs), which drive chemo- and radio-resistance. Statins, known to inhibit CRC progression, have been suggested to enhance chemosensitivity by targeting TICs, though their exact mechanism remains unclear. We have also observed altered SATB1/2 levels in patient tumors compared to normal tissues. This study aims to establish SATB proteins as diagnostic and therapeutic targets, exploring their genetic and epigenetic impact using CRC cell lines, patient samples, organoids, and xenograft models.
Keywords: Colorectal cancer, Cancer stem cells, Colonoid, statins, adjuvant therapy, drug repurposing
Bullet points (highlights)
- Role of SATB family chromatin organizer proteins in CRC development and progression and cellular plasticity
- Delineate the role of statins in regulating SATB proteins and cancer stem cells
- Repurposing statins as therapeutics for CRC patients
- Study how SATB proteins regulate gene expression, tumor dynamics, and cancer progression in colorectal cancer using colon organoids
Graphical abstract
Active collaborations:
- Drs Prachi Patil and Avnish Saklani: Tata Memorial Hospital, Mumbai for randomized phase 2/3 clinical trial evaluating the therapeutic potential of statin as antitumor agent in CRC.
- Dr Sorab Dalal: ACTREC, Kharghar: for studying CRC patient derived organoids.
- Prof. Tae-Young Roh: Laboratory of System Genomics, EWHA Womans University, Seoul: for generating colon organoids and knockout models.
Blurb:
- Statins as antitumor agents
SATB proteins function as downstream effectors in Wnt/β-catenin signaling. Our research has demonstrated that statins influence SATB protein levels in CRC cells. Our study aims to elucidate the mechanistic regulation of SATB protein expression following statin treatment in CRC cell lines, mice, and CRC patients enrolled in statin clinical trials. We propose that the combined expression patterns of SATB1/2 may serve as potential biomarkers for CRC prognosis.
- Cancer stem cells (CSC) in CRC
Disruptions in intestinal stem cell homeostasis and Wnt signaling are often linked to colorectal cancer (CRC) development. Our research aims to investigate the role of Wnt effector-SATB proteins in regulating cellular plasticity in CRC, utilizing CRC cell lines, spheroids, orthotopic tumor implants, and PDX and PDO models.
- Colonoids as a model to study CRC
Colon organoids serve as a robust 3D model for studying colorectal cancer progression as they effectively replicate the hierarchical structure of the intestine. Additionally, this model enables the exploration of SATB1/2's role in gene expression, cell differentiation, and tumor dynamics in colon cancer. By closely mimicking the in vivo tumor microenvironment, it provides a valuable platform to investigate the distinct functions of SATB1/2 in cancer development and progression.
Publications:
- Reddy CNL, Vaijayanti VN, Notani D, Galande S, Kotamraju S. Down-regulation of the global regulator SATB1 by statins in COLO205 colon cancer cells. Med. Rep. 2010; 3:857-861.
- Mir R, Pradhan SJ, Galande S. Chromatin organizer SATB1 as a novel molecular target for cancer therapy. Curr Drug Targets. 2012; 13:1603-1615.
- Mir R, Pradhan SJ, Patil P, Mulherkar R and Galande S. Wnt/β-catenin signalling regulated SATB1 promotes colorectal cancer tumorigenesis and progression. Oncogene. 2015; 35:1679-1691.
- Naik R, and Galande S. SATB family chromatin organizers as master regulators of tumor progression. Oncogene2019; 38: 1989-2004.
- Tripathi S, Gupta E, and Galande S. Statins as anti‐tumor agents: A paradigm for repurposed drugs. Cancer Reports2024; 7: e2078.
- Tripathi S, et al. Statins attenuate Wnt/β-catenin signaling by targeting SATB family proteins in colorectal cancer. bioRxiv(2024): 08.23.609189; doi: https://doi.org/10.1101/2024.08.23.609189
Colin Jamora:
Summary: Our laboratory focuses on elucidating the mechanisms underlying diseases that arise from dysregulation of the wound healing program in the skin. Wound healing is a tightly orchestrated process comprising three distinct but overlapping phases: inflammation, proliferation, and remodeling. Disruptions in these phases contribute to the pathogenesis of several common and debilitating diseases. Our research aims to dissect the cellular interactions and signaling pathways perturbed in these conditions, aiming to translate these insights into novel therapeutic strategies.
Our areas of focus include:
- Inflammatory skin disorders: Investigating the molecular basis of conditions such as psoriasis and eczema.
- Impaired wound healing in diabetes: Understanding the barriers to efficient skin repair in diabetic contexts.
- Tissue fibrosis: Exploring fibrotic processes, particularly in the skin, as observed in diseases such as scleroderma.
- Cutaneous malignancies: Examining the role of wound healing deregulation in skin cancer pathogenesis.
Graphical Abstract:
from doi: 10.1016/j.celrep.2022.111390
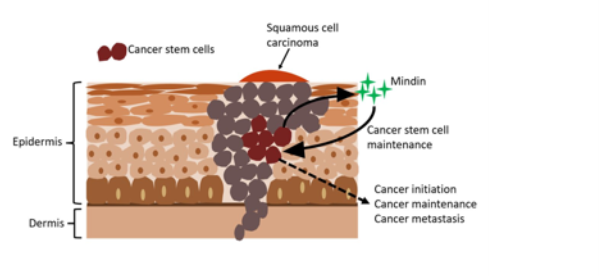
Tumors consist of various cell types, with cancer stem cells playing a crucial role in driving tumor growth and recurrence. These specialized cells possess unique abilities: self-renewal, which enables them to generate additional cancer stem cells, and differentiation, allowing the formation of other tumor cell types. These properties are central to cancer initiation, progression, and maintenance. Furthermore, cancer stem cells often exhibit resistance to conventional therapies, contributing to relapse and metastasis. A promising approach to achieving long-lasting tumor elimination while reducing the risk of recurrence is specifically targeting these cells.
Using a genetically engineered mouse model, the Jamora lab identified a mechanism involving the protein Mindin (also known as Spondin-2), which is crucial in maintaining cancer stem cells. Remarkably, their findings revealed that inhibiting Mindin disrupts cancer stem cell function and effectively halts tumor growth. These insights suggest that targeting Mindin could offer a highly effective therapeutic strategy to prevent cancer relapse and recurrence.
Active collaborations:
- KoshKey Sciences Pvt. Ltd (Bangalore).: Diabetic wound healing
- PhytoKosh (Bangalore): Development of natural products with anti-viral and pro-healing activity
- Vikas Agrawal, MD (Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow): Fibrosis and Scleroderma
- John Varga, MD (University of Michigan, USA): Fibrosis and Scleroderma
- Manjunath Joshi, PhD (Manipal Academy of Higher Education, Manipal): Diabetes and Inflammation
- Vivek Malhotra, PhD (Centre for Genomic Regulation, Barcelona): Fibrosis therapies
Sri Krishna Jayadev Magani
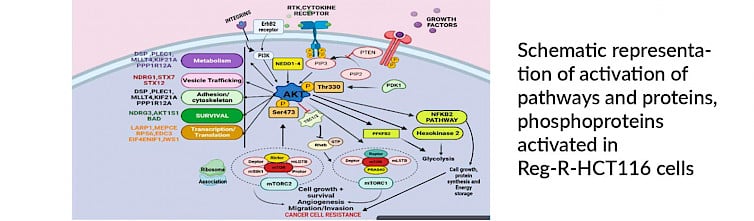
Summary: Acquired drug resistance is one of the major factors for the recurrence and lethality in all cancers, including colorectal cancer. Kinases are reported to play a major role in signalling cell survival and proliferation of cancer cells. Kinase inhibitors targeting multiple kinases are currently being used as one of the therapeutic strategies for treating cancers. With recent reports of resistance to Sorafenib and regorafenib, understanding the mechanism of acquired resistance development to these drugs is important for proper patient management. My labs research interest is in understanding the cancer cell signalling in disease progression and therapy. Presently, we are working on understanding the mechanism of drug resistance development to multi-kinase inhibitor drugs in colorectal cancers. For this we have developed a regorafenib resistant HCT116 cell line (Reg-R-HCT116) and are using multi-omics approach to understand molecular signalling alteration leading to the development of drug resistance in these cells. The proteome, phospho-proteome, exome and methylome data generated hints about some novel molecules and different cell organelle in resistance development.
Along with mechanistic understanding of the resistance development we are also interested in development and screening of novel anticancer agents . The major type of molecules of this study are either natural compounds or organometallic compound. In organometallic compound we are working with ferrocene and ruthenium based compounds and are also trying to understand their mechanism of action. Our other interests include understanding the signalling in some rare tropical diseases like tropical calcific pancreatitis.
Highlights:
- Development of regorafenib resistant HCT116 cell line.
- Proteome, phospho-proteome, exome and methylome data generation
- To understand the role of PI3K/AKT/ mTOR and TGFb signalling in drug resistance.
- The role of Mitochondria, ribosomes and ER in resistance development.
- To understand the metabolic changes and their role in resistance development
- Screening of ruthenium arene compounds and ferrocene ruthenium compounds for their anticancer activity.
Graphical abstract:
Active collaborations:
- Drs Ashutosh and Dr. P C Reddy in the department are helping us in the omics data analysis.
- Dr Dasaradhi from inSTEM Bangalore is helping us in evaluating the role of cell organelle in resistance development and disease progression.
- Dr Raj kumar Joshi from MNIT Jaipur is a collaborator helping us and supplying the organometallic compound for the anticancer drug screening.
Drs Srinivasan from Tribal University, Andhra Pradesh and Dr. Shanmukha Anand from GITAM university Visakhapatnam are collaborating with us in the Natural compound screening.
Richa Priyadarshini
Summary: The bacterial cell wall comprises peptidoglycan (PG), which consists of alternating N-acetylglucosamine and N-acetylmuramic acid residue cross-linked by peptide side chains. Far from being static, the cell wall is constantly remodelled to allow cell growth and expansion. Maintaining the integrity of the cell wall is essential for cell viability, and its importance is evident by the number of antibiotics that target the PG metabolic pathway, such as the ß-lactam antibiotics. Enzymes that cleave specific bonds in the PG are ubiquitous among bacteria. PG cleaving enzymes are collectively known as PG hydrolases or cell wall-modifying enzymes and can be further classified into different groups depending on the type of bonds they cleave. Amidases remove the peptide side chain, lytic transglycosylases hydrolyse the glycan backbone, and endopeptidases cleave the peptide cross-links. The precise physiological roles for some of these enzymes remains obscure till now due to redundancy in bacterial cells
While it is generally believed that beta-lactam treatment leads to loss of cell wall integrity, very little is known about the mechanism of action of these drugs. Recent studies probing the mechanism of action of beta-lactams have revealed a prominent role for PG hydrolases. Even though much is known about the enzymatic activity and structure of cell wall-modifying enzymes, their regulation in the bacterial cell is not elucidated. Many questions regarding their spatial and temporal regulation remain unanswered. How is the activity of these cell wall-modifying enzymes coordinated with PG synthesis in time and space to ensure that the integrity of the cell wall remains intact at all times? Our lab uses two well-studied bacterial model systems E.coli and Caulobacter crescentus, to study cell wall biogenesis. Insights into regulating cell wall-modifying enzymes will close a fundamental gap in our understanding of bacterial cell morphogenesis.
Apart from this we are testing material and chemical compounds for antibacterial activity in collaborative projects
Highlights:
- Understand the physiological roles PG modifying enzymes in bacterial cell growth and division
- Spatiotemporal regulation of PG enzymes in cells
- Role of PG modifying enzymes in antibiotic stress.
Funding:
SERB-CRG, CSIR-EMR
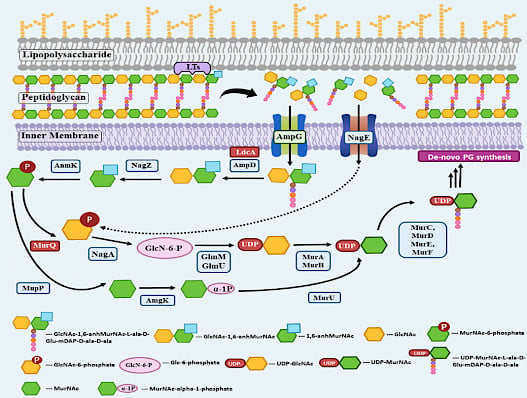
Peptidoglycan recycling pathway in Caulobacter crescentus. Modi et al. 2025. mBio
Publications:
- Malvika Modi, Deepika Chauhan, Michael C. Gilmore, Felipe Cava and Richa Priyadarshini. Deficiency in peptidoglycan recycling promotes β-lactam sensitivity in Caulobacter crescentus. 2025. Feb. mBio
- Rath S, Maiti D, Modi M, Pal P, Munan S, Mohanty B, Bhatia A, Bhowal R, Priyadarshini R, Samanta A, Munshi P, Sen S. Metal-free synthesis and study of glycine betaine derivatives in water for antimicrobial and anticancer applications. iScience. 2023 Jul 13;26(8):107285. doi: 10.1016/j.isci.2023.107285. PMID: 37575199; PMCID: PMC10415718.
- Sangeeta Sahu, Rashmi Niranjan , Richa Priyadarshini *, Bimlesh Lochab* Benzoxazine-grafted-chitosan biopolymer films with inherent disulfide linkage: Antimicrobial properties. Chemosphere. 2023 Jul;328:138587. doi: 10.1016/j.chemosphere.2023.138587. Epub 2023 Apr 3. PMID: 37019400.
- Paramita Sarkar, Kathakali De, Malvika Modi, Geetika Dhanda, Richa Priyadarshini, Julia E. Bandow, Jayanta Haldar* Next-generation membrane-active glycopeptide antibiotics that also inhibit bacterial cell division. 2023. Sci., DOI :https://doi.org/10.1039/D2SC05600C
- Rashmi Niranjan , Saad Zafar,Bimlesh Lochab* , Richa Priyadarshini* Synthesis and Characterization of Sulfur and Sulfur-Selenium Nanoparticles Loaded on Reduced Graphene Oxide and Their Antibacterial Activity against Gram-Positive Pathogens. Nanomaterials (Basel). 12(2):191.
- Deepika Chauhan, Santanu Hati, Richa Priyadarshini*, Subhabrata Sen*. Transcriptome analysis predicts mode of action of benzimidazole molecules against Staphylococcus aureus UAMS‐1. Drug Dev Res. 1–14.
- Amrita Dubey and Richa Priyadarshini. Amidase activity is essential for medial localization of AmiC in Caulobacter crescentus. 2017. Current Genetics 64 (3), 661-675.
Naga Suresh Veerapu
Major Research Areas:
- Understanding the life cycle of single-strand positive sense RNA viruses.
- Developing cell culture systems for viruses to study viral genetic diversity and host-virus interactions.
- Identifying novel antivirals against public health concern viruses.
Publications:
- Shaheen Khan, Yashwant Kumar, Charu Sharma, Sonu Kumar Gupta, Amit Goel, Rakesh Aggarwal, Naga Suresh Veerapu. Dysregulated Metabolites and Lipids in Serum of Patients with Acute Hepatitis E: A Longitudinal Study, Journal of Viral Hepatitis, 2023.
- Soni S, Agarwal S, Aggarwal R, Veerapu NS. Reverse Genetics to Engineer Positive-sense RNA Virus Variants. J Visual Experiments, 2022.
- Soni S, Singh D, Aggarwal R, Veerapu NS. Enhanced Replication Capacity of Hepatitis C Virus Increases Resistance to Direct-Acting Antivirals. Journal of General Virology, 2022.
- Khan S, Soni S, Veerapu NS. HCV Replicon Systems: Workhorses of Drug Discovery and Resistance. Frontiers Cellular and Infection Microbiology, June 2020.
- Singh D, Soni S, Khan S, Sarangi AN, Yennamalli RM, Aggarwal R, Veerapu NS. Genome-wide mutagenesis of hepatitis C virus reveals genome ability to overcome detrimental mutations. Journal of Virology, 2020.
- Agarwal S, Baccam P, Aggarwal R, Veerapu NS. Novel Synthesis and Phenotypic Analysis of Mutant Clouds for Hepatitis E Virus Genotype 1. Journal of Virology, 2018.
Virus culture facility

Ashish Gupta:
Summary: My lab is dedicated to advancing the understanding of prostate cancer, focusing on how the androgen receptor (AR) is deregulated by epigenetic factors. Prostate cancer is one of the leading causes of cancer-related deaths, and its progression is closely tied to AR activity. This critical transcription factor drives tumor growth and survival. While androgen deprivation therapy (ADT) is initially effective, many prostate cancers eventually develop resistance, resulting in the emergence of castration-resistant prostate cancer (CRPC). This resistance is often associated with AR dysregulation, even in low androgen conditions. We aim to investigate the complex interplay between AR and various epigenetic modifications that contribute to its aberrant activation and signaling. By understanding these epigenetic alterations, we seek to identify novel therapeutic targets and develop strategies to combat AR-driven prostate cancer, particularly in cases where traditional treatments fail. Through our work, we hope to uncover vital insights into the molecular mechanisms driving prostate cancer and contribute to developing more effective, targeted treatments that can improve patient outcomes.
Graphical abstract
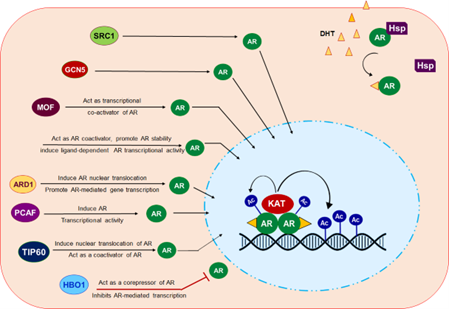
From: Jaiswal B et al (2022) Front. Endocrinol. 13:886594.
Focus research area-
- Understanding deregulation of AR by epigenetics modifier TIP60
- screening and identification of new generation drugs to target AR-TIP60 complex
Anindita Chakrabarty:
Summary: Cancer is a leading non-communicable disease, with alarming statistics that demand our attention. In 2023, the WHO estimated around 20 million newly diagnosed cancer cases and approximately 10 million cancer-related deaths. These figures are projected to rise by over 60% in the next two decades, underscoring an urgent health crisis. As cancer predominantly affects the elderly, the global rise in life expectancy significantly contributes to this escalating burden.
We focus on two research areas to combat rising cancer rates and mortality. First, we study the impact of inflammation on the modification of reactive cysteine residues in DNA repair proteins, which could shed light on inflammation-associated carcinogenesis. Second, we investigate treatment-induced senescence and its role in drug resistance, which affects relapse and mortality. We also explore how insulin-like growth factor (IGF) signaling contributes to tumor relapse by neutralizing reactive oxygen species and promoting tumor neurogenesis. Our goal is to use IGF1 signaling inhibitors to enhance the efficacy of cancer chemotherapy.
Together, these research avenues represent a crucial step toward understanding and ultimately reducing the impact of cancer on our society. It is vital that we invest in these studies to pave the way for innovative treatments and hope for those affected by this devastating disease.
Keywords: Inflammation, Carcinogenesis, Cysteine Modification, Therapy Induced senescence, Drug resistance, Evolutionary therapy, Breast Cancer, Natural Products
Highlights of the current projects:
- Formation of DNA-protein cross-link formation by DNA repair proteins such as AGT/MGMT at the site of inflammation by forming cross-link with 8-nitro-guanine DNA lesion
- Maladaptive effects of inflammation-associated DNA-protein cross-links on innate immunity and cancer predisposition
- Dynamic emergence of non-genetic drug tolerance through therapy-induced senescence and adaptive strategies to prevent it
- 4. IGF-1 in redox homeostasis in cancer cells and cells of tumor microenvironment
- Repurposing different anticancer natural products as broad-spectrum antivirals
Relevant Publications:
- Shayantani Chakraborty, Shaista Haider, Gargi Mukherjee, Anindita Chakrabarty*, and Goutam Chowdhury*. O6-Alkylguanine-DNA Alkyltransferase Maintains Genomic Integrity by Forming DNA-Protein Crosslinks During Inflammation-associated Peroxynitrite-Mediated DNA Damage. ACS Chem Res Toxicol 2024, 37(12): 1952-1964
- Shaista Haider, Shayantani Chakraborty, Goutam Chowdhury*, Anindita Chakrabarty*. Opposing Interplay between Nuclear Factor Erythroid 2-Related Factor 2 and Forkhead BoxO 1/3 is Responsible for Sepantronium Bromide’s Poor Efficacy and Resistance in Cancer cells: Opportunity for Combination Therapy in Triple Negative Breast Cancer. ACS Pharmacology & Translational Science, 2024, 7(5), 1237-1251
- Shayantani Chakraborty, Dibyendu Mallick, Mausumi Goswami, F. Peter Guengerich, Anindita Chakrabarty*, and Goutam Chowdhury*. The Natural Products Withaferin A and Withanone From the Medicinal Herb Withania somnifera are Covalent Inhibitors of the SARS-CoV-2 Main Protease. ACS J Nat. Prod. 2022, 85(10), 2340-2350 (Front cover page article)
- Anindita Chakrabarty*, Shayantani Chakraborty, Ranjini Bhattacharya, Goutam Chowdhury. Senescence-induced chemoresistance in triple-negative breast cancer and evolution-based treatment strategies. Front. Oncol., 2021, 674354 (Invited review)
5. Nabeel Ahmed, Anindita Chakrabarty, F. Peter Guengerich, Goutam Chowdhury. Protective Role of Glutathione against Peroxynitrite-Mediated DNA Damage During Acute Inflammation. ACS Chem. Res. Toxicol. 2020, 33(10) 2668-2674
Graphical Abstracts:
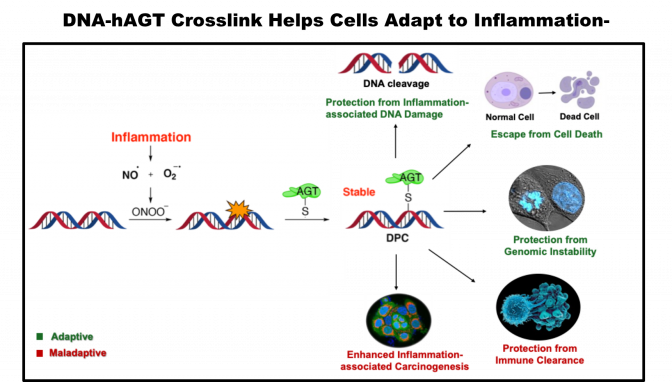
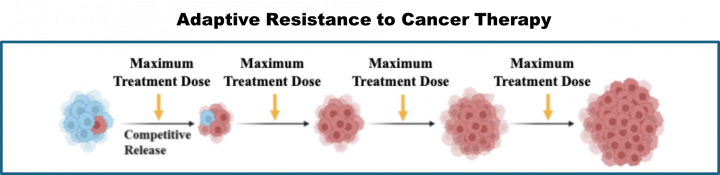
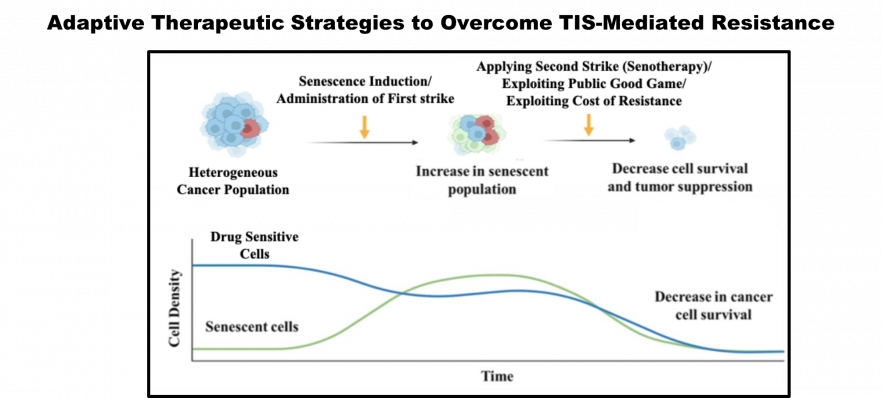
Active Collaborations:
Dr. F. P. Guengerich: Department of Biochemistry, Vanderbilt University Medical Center, USA
Dr. Goutam Chowdhury: Institut de R&D Servier Paris-Saclay, Gif-sur-Yvette, France
Dr. Imtaiyaz Hassan: Center for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, Delhi
Dr. Bani Kanta Sarma: Jawaharlal Nehru Center for Advanced Scientific Research, Bangaluru
Dr. Satendra Kumar: Department of Surgery, Government Institute of Medical Sciences, Greater Noida
Dr. Manisha Singh: Department of Biochemistry, Government Institute of Medical Sciences, Greater Noida
Rajan Vyas
Summary: Glycosaminoglycans (GAGs) are heteropolysaccharides primarily located on the animal cell surface in the extracellular matrix (ECM) and known to bind and regulate several proteins such as chemokines, growth factors, enzymes, cytokines, and adhesion proteins involved in physiological and pathological processes. GAGS are formed of repeating disaccharide units of amino sugars (N-acetylglucosamine [GlcNAc] or N-acetylgalactosamine [GalNAc]) and either hexuronic acid or hexose. GAGs are of two types: Non-sulfated GAGs, which include hyaluronic acid (HA), while sulfated GAGs comprise chondroitin sulfate (CS), dermatan sulfate (DS), keratan sulfate (KS), heparin and heparan sulfate (HS). CS consists of alternating β (1–4)-linked GalNAc and β (1–3)-linked glucuronic acid (GlcUA) disaccharide units exhibiting sulfate groups at various positions, i.e., sulfation at the C-4 position of GalNAc forms CS-A, sulfation at C-6 position of GalNAc forms CS-C and sulfation at both C4 and C6 positions of GalNAc results in the formation of CS-E. These sulfated CS are linked to proteins as part of a proteoglycan and are involved in various crucial biological processes, including mammalian chondrocyte development and growth factor signaling during embryonic cartilage morphogenesis. Moreover, along with development, these enzymes have also been reported to be essential and tightly regulated during injury and disease. Surprisingly, changes in sulfation balance and pattern of sulfation on the glycoproteins surface have been correlated with enhancing the metastatic efficiency of tumor cells and affecting cancer cell behavior via MAPK signaling pathways involved in cancer progression. Hence, it is critical to understand how sulfotransferases are regulated for proper sulfation on CS chains.
Highlights:
- Enzymes controlling the sulfation of CS are essential for various biological processes and play a crucial role in development, signaling, and disease.
- Sulfotransferase enzymes that control the sulfation of CS could be used as a potential target for drug/preclinical and clinical therapy.
Graphical abstract:
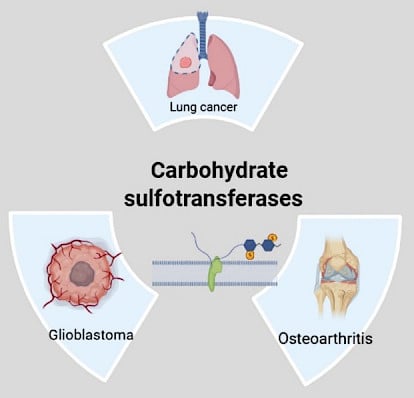
Active collaboration:
- Banaras Hindu University (BHU), Varanasi
Relevant facilities:
- Protein expression and purification facility
- Protein analysis facility – Circular dichroism (CD) spectrophotometer, Microscale thermophoresis (MST)