Development, Regeneration and Repair
Development, Regeneration and Repair
We focus on cellular and molecular principles governing the important processes for tissue, organ and organism development, regeneration and repair. We study these in multiple invertebrate, vertebrate and in vitro systems. model systems, as well as in organisms ranging from Hydra to mammalian models and utilize our state-of-the-art facilities for animal research (CITRes, Fly facility), Epigenetics Research (COEE), Imaging (Imaging facility), NGS and mass spectrometry.
Research Groups
Colin Jamora

Our laboratory aims to uncover the cellular and molecular mechanisms of tissue regeneration and repair for therapeutic applications. We use genetic engineering, cell biology, and quantitative biology to study tissue homeostasis, with a focus on skin. As one of the few tissues that regenerates cyclically throughout life, the skin epithelium also has a remarkable ability to rapidly repair itself after injury. Using this as our experimental paradigm, our research focuses on two key areas: (1) the early cellular events that trigger wound healing and transition epidermal keratinocytes from homeostasis to repair, and (2) the interactions between the extracellular matrix, vasculature, inflammatory cells, and fibroblasts that drive scar formation and fibrosis. These studies provide insight into diseases linked to wound-healing mechanisms, reinforcing the “wound signature” hypothesis. While rooted in fundamental skin biology, our work also has strong translational potential, fostering collaborations with the pharmaceutical and biotech industries to develop novel therapies.
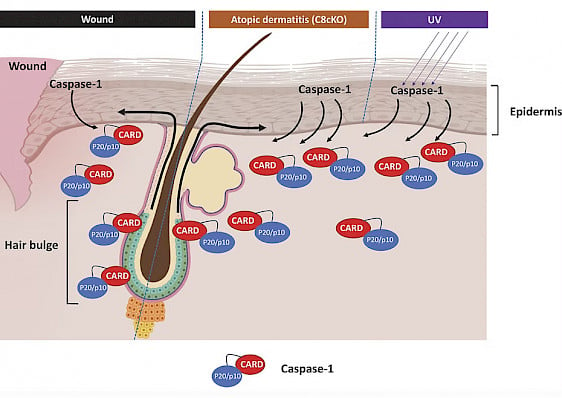
Model of Caspase-1 mediated HFSC homing to the epidermis upon wounding. Stressed epidermal keratinocytes secret Caspase-1 to the extracellular milieu upon wounding, UV exposure or genetic ablation of Caspase-8. Via caspase activation and recruitment domain (CARD) of caspase-1, hair follicle stem cells are coaxed to migrate to the epidermis. (from Hegde et al., 2024).
Projects/Highlights
-
Non-immune Functions of Immune Cells: Investigating the unconventional roles of immune cells in regulating resident skin cells during wound healing and cutaneous regeneration.
-
Stem Cell Regulation in Tissue Repair: Exploring the mechanisms governing stem cell activity in skin homeostasis and the tissue repair process.
-
Cell Fate Transitions: Deciphering the signaling pathways that drive cellular transitions critical to regeneration and healing.
-
Mechanobiology and Epigenetics: Examining the influence of mechanobiological forces and epigenetic modifications on the initiation and progression of wound healing.
Key Publications
-
Hegde, A., Ghosh, S., Ananthan, A. S., Kataria, S., Dutta, A., Prabhu, S. et al. (2024) Extracellular Caspase-1 induces hair stem cell migration in wounded and inflamed skin conditions J Cell Biol 223, 10.1083/jcb.202306028
-
Dutta, A., Saha, D., and Jamora, C. (2024) Approaches to Study Wound-Induced Hair Neogenesis (WIHN) Methods Mol Biol 10.1007/7651_2024_522
-
Bhatt, T., Dey, R., Hegde, A., Ketkar, A. A., Pulianmackal, A. J., Deb, A. P. et al. (2022) Initiation of wound healing is regulated by the convergence of mechanical and epigenetic cues PLoS Biol 20, e3001777 10.1371/journal.pbio.3001777
-
Badarinath, K., Dam, B., Kataria, S., Zirmire, R. K., Dey, R., Kansagara, G. et al. (2022) Snail maintains the stem/progenitor state of skin epithelial cells and carcinomas through the autocrine effect of matricellular protein Mindin Cell Rep 40, 111390 10.1016/j.celrep.2022.111390
Kamalesh Kumari

Our research focuses on understanding the cellular and molecular mechanisms of biological membrane remodeling, particularly in the context of exocrine vesicle exocytosis. Exocrine glands play a crucial role in human physiology by secreting digestive enzymes, surfactants, mucus, and hormones onto epithelial surfaces to regulate cell signaling, metabolism, and immune defense functions. Exocrine secretion relies on giant, micron-sized vesicles, which fuse with the apical surface of cells upon stimulation. Traditionally, this process was believed to occur via full-collapse exocytosis, but such a mechanism would add tremendous amounts of membranes to the cell surface, disrupting surface homeostasis in terms of size, shape, and composition. Using live-cell super-resolution imaging, correlative light and electron microscopy (CLEM), and biophysical assays in Drosophila larval salivary glands and mouse acinar pancreases ex vivo primary culture models, we uncovered a novel pathway of exocytosis. We found that, after fusion, vesicle membranes undergo dynamic remodeling and crumpling, ensuring mechanical sequestration of the vesicular membrane while allowing complete cargo extrusion—without apical surface expansion. We refer to this as Membrane Crumpling-mediated Exocytosis (Mem-CruX). Using live-imaging and quantitative analysis with biochemistry, our research aims to identify key molecular remodelers involved in the Mem-CruX process and investigate how errors in membrane remodeling contribute to secretory dysfunctions and disease states (like pancreatic and pulmonary insufficiencies), providing insights for targeted therapeutic interventions in exocrine disorders.
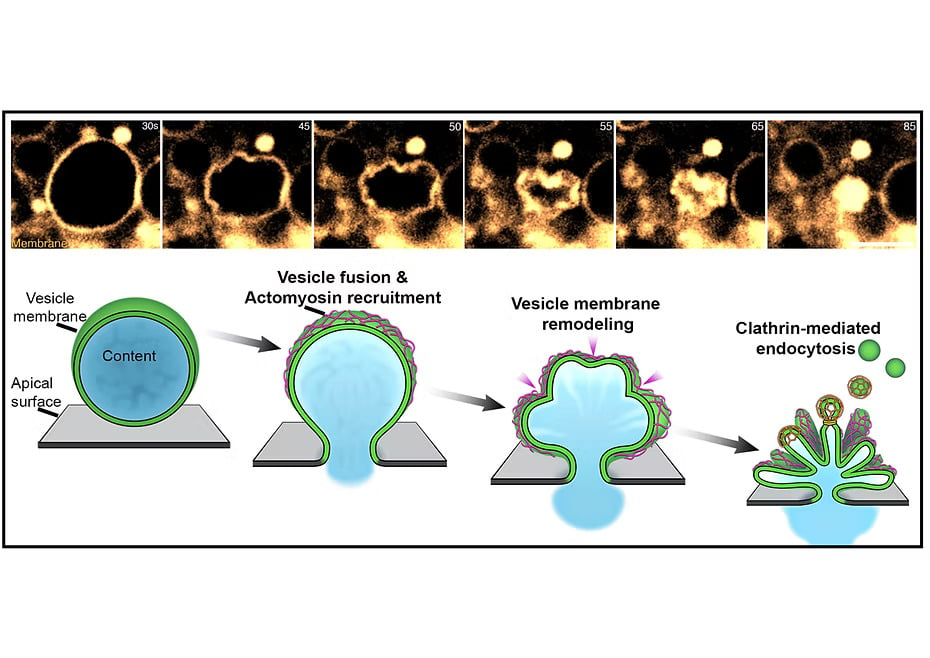 Top Panel: Live imaging of vesicular secretion captured using Airy Scan super-resolution microscopy, showing the membrane crumpling process during exocytosis.
Top Panel: Live imaging of vesicular secretion captured using Airy Scan super-resolution microscopy, showing the membrane crumpling process during exocytosis.
Bottom Panel: Schematic representation of the Membrane Crumpling-Mediated Exocytosis (Mem-CruX) pathway. Following vesicle fusion with the apical surface, a diffusion barrier is established at the interface, preserving the physicochemical properties of the apical membrane while keeping the vesicle membrane distinct. Next, actomyosin machinery is recruited to the fused vesicle membrane, initiating progressive crumpling and folding. This enables efficient cargo extrusion while mechanically sequestering the vesicle membrane, preventing apical surface expansion. Subsequent to cargo release, the crumpled vesicle membrane undergoes targeted endocytosis via the clathrin-mediated pathway, ensuring membrane recycling and homeostasis. While the mechanical and structural framework of Mem-CruX is emerging, the molecular players orchestrating this process remain unexplored, representing a key focus of our ongoing research.
Projects/Highlights
-
Live-Imaging Candidate Screen & Characterization – Identify and analyze membrane remodelers involved in Mem-CruX using the Drosophila 3rd instar larval salivary gland model.
-
Role of Lipid Regulators in Mem-CruX – Investigate how lipids and their modifying enzymes contribute to vesicle membrane crumpling and homeostasis.
-
Biochemical Isolation & Molecular Characterization of Secretory Vesicles – Purify and analyze secreting vesicles from mouse pancreatic acinar cells to identify key molecular components of Mem-CruX.
-
Fate of Endocytosed Vesicular Membrane – Determine how endocytosed vesicle membranes are processed and recycled following Mem-CruX.
-
Biophysical Properties of the Apical Surface & Vesicle Membrane – Characterize the mechanical and physicochemical properties of the apical membrane, vesicle membrane, and cargo regulating Mem-CruX.
-
Impact of Inflammation & Injury on Mem-CruX Dynamics – Examine how inflammatory signals and cellular injury influence Mem-CruX in pancreatic acinar cells and lung alveoli.
Key Publications
-
Exocytosis by vesicle crumpling maintains apical membrane homeostasis during exocrine secretion. K. Kamalesh, N. Scher, T. Biton, E. D. Schejter, B. Z. Shilo, O. Avinoam, Dev. Cell. 56, 1603-1616 (2021). DOI: 10.1016/j.devcel.2021.05.004· Highlighted article: A mechanochemical mechanism couples exocrine secretion to endocytic membrane retrieval. V. Haucke, Dev. Cell. 56, 1557-1559 (2021).
-
Kamalesh, K., Segal, D., Avinoam, O., Schejter, E.D., and Shilo, B.-Z. Structured RhoGEF recruitment drives myosin II organization on large exocytotic vesicles. J Cell Sci. 2024 Jul 1;137(13):jcs261944.
Puli Chandramouli Reddy

Organisms have evolved with diverse cell types and body plans that facilitate specialization and coordination. While cell types segregate functions, body plans organize them to ensure efficient, integrated activity. Morphological diversity reflects the various solutions organisms have adopted for survival in diverse environments. Understanding the molecular mechanisms underlying the evolution of cell types and tissue organization is a fundamental question in biology. This knowledge is crucial for elucidating how diseases or disorders arise from cellular dysfunction and misassembly during development. It also provides insights into unique processes such as regeneration and the emergence of specialized cell types and functions that enhance survival. Addressing these questions is the long-term focus of my research.
Early-branching basal metazoans with simple well-defined body plans and cell types comparable to those in bilaterians offer powerful models for exploring these evolutionary questions. Hydra, a member of the phylum Cnidaria, is particularly valuable due to its simple oral-aboral axis, the emergence of key cell types such as neurons and muscle cells, and its exceptional regenerative capacity. Hydra’s remarkable tissue dynamics enable continuous axis patterning, making it an ideal system for investigating the molecular networks driving body axis development and regeneration.
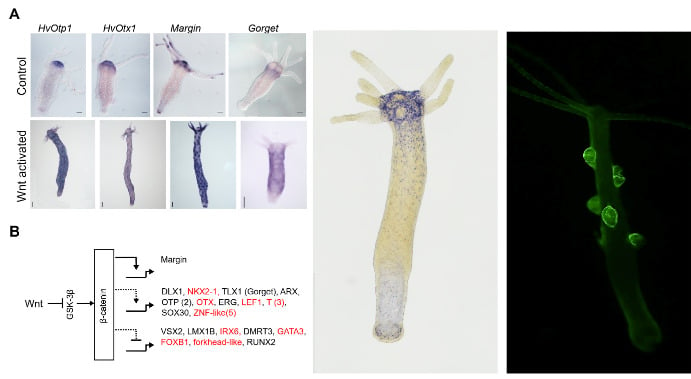
Left Panel: Conserved and newly identified homologs of bilaterian genes involved in axis patterning and regulated by the Wnt/β-catenin signaling pathway in Hydra. (modified from Reddy et al., 2019)
Middle Panel: Expression pattern of a novel neuropeptide identified in the lab. (unpublished)
Right Panel: Transgenic reporter line showing GFP expression driven by a gene that express in somatic cells of the male gonad in Hydra. (unpublished)
Projects/Highlights
- Signaling and epigenetic regulation during Hydra regeneration
- Chromatin dynamics during germ-cell development in Hydra
- Interaction of microbiome and epigenome in Hydra
- Insights from the Secretome: Exploring the dynamics of secretory proteins and small peptides in regeneration and the evolution of cell-cell communication from a peptidergic signaling perspective.
Key Publications
- Reddy, P.C., Gungi, A., Ubhe, S., and Galande, S. (2020). Epigenomic landscape of enhancer elements during Hydra head organizer formation. Epigenetics & Chromatin 13, 1-16.
- Reddy, P.C., Gungi, A., Ubhe, S., Pradhan, S.J., Kolte, A., and Galande, S. (2019a). Molecular signature of an ancient organizer regulated by Wnt/β-catenin signalling during primary body axis patterning in Hydra. Communications biology 2, 434.
- Reddy, P.C., Gungi, A., and Unni, M. (2019b). Cellular and molecular mechanisms of Hydra regeneration. Evo-Devo: Non-model Species in Cell and Developmental Biology, 259-290.
- Reddy, P.C., Ubhe, S., Sirwani, N., Lohokare, R., and Galande, S. (2017). Rapid divergence of histones in Hydrozoa (Cnidaria) and evolution of a novel histone involved in DNA damage response in hydra. Zoology 123, 53-63.
- Reddy, P.C., Unni, M.K., Gungi, A., Agarwal, P., and Galande, S. (2015). Evolution of Hox-like genes in Cnidaria: Study of Hydra Hox repertoire reveals tailor-made Hox-code for Cnidarians. Mechanisms of development 138, 87-96.
All publications link: https://scholar.google.com/citations?hl=en&user=LPPmAyoAAAAJ&view_op=list_works&authuser=1&sortby=pubdate
Rohini Garg

Our lab tries to understand the role of G-quadruplexes and its associated helicases in plant development. In addition, we are also trying to understand the effect of gene regulatory regions on plant root, flower and seed development. We are initiated work on understanding the role of various non-coding RNAs and small peptides in plant regeneration and root development.
Projects/Highlights
- Role of GQS helicases in plant development
- Effect of small peptides in plant regeneration
- Role of GQSes in gene regulation during plant development
- Role of non-coding RNAs in root development
Key Publications
- H Sharma & Garg R (2025). Knot-Knot Chronicles: Unveiling the G-Quadruplexes. Critical Reviews in Biotechnology (accepted). https://doi.org/10.1080/07388551.2024.2435960. (IF=8.2)
- Ali N, Singh S and Garg R (2025). Unlocking Crops' Genetic Potential: Advances in Genome and Epigenome Editing of Regulatory Regions. Current opinion in Plant Biology (accepted). 83, 102669 https://doi.org/10.1016/j.pbi.2024.102669. (IF = 8.3)
- Garg R., Sahu SK, Jain M, (2024) Single same-cell multiome for dissecting key plant traits. Trends in Plant Sciences, online DOI: 10.1016/j.tplants.2024.10.008. Spotlight article (IF = 17.3)
- Rajkumar MS, Tembhare K, Garg R, Jain, M. (2024) Genome-wide mapping of DNase I hypersensitive sites revealed differential chromatin accessibility and regulatory DNA elements under drought stress in rice cultivars. The Plant Journal : for cell and molecular biology, 119(4), 2063–2079. https://doi.org/10.1111/tpj.16864
- MS Rajkumar, K Gupta, NK Khemka, R Garg†, M Jain. (2020). DNA methylation reprogramming during seed development and its functional relevance in seed size/weight determination in chickpea. Communications biology 3 (1), 1-13. (†Joint corresponding author) (IF = 5.9)
- N Khemka, M Singh Rajkumar, R Garg, M Jain (2021). Genome‐wide profiling of miRNAs during seed development reveals their functional relevance in seed size/weight determination in chickpea. Plant direct, 5 (3), e00299. (IF = 3.0)
- Garg R*, Aggarwal J, Thakkar B. (2016). Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Scientific Reports 6, 28211. (*corresponding author). (IF = 4.6).
Rudra Nayan Das

Vascular networks, the lymphatic vessels (LV) and the blood vessels (BV), in the vertebrate body display an extraordinary range of morphological and functional diversity. These complex LV/BV systems, composed of endothelial cells (ECs), engage in close interactions with diverse organ microenvironments. Our focus lies in unraveling how ECs develop distinct vessel and organ-specific identities and understanding their vital roles in organ development, function and regeneration.
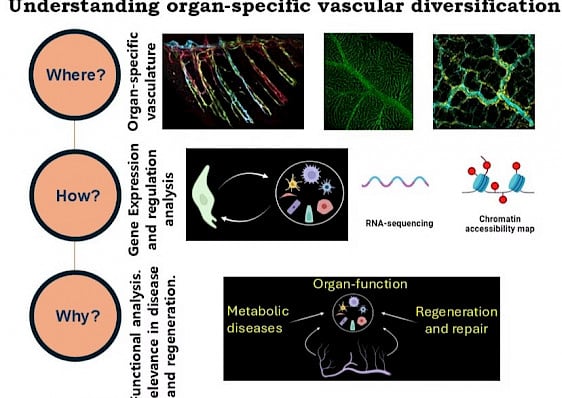
We investigate organ-associated vascular diversity through three broad question: Where, why and how? In the where questions, we characterize the vascular populations in different organs through confocal microscopy and RNA sequencing. In the how questions, we investigate the molecular mechanisms underlying these organ-associated vascular specializations. In the why questions, we explore the functional aspects of these specializations and focus on how these are associated with various vasculopathies or regeneration mechanism.
Projects/Highlights
-
Regulation of Lymphatic Vessel specialization and gene regulation: We aim to understand how lymphatic vessels develop in different organs and acquire organ-specific specializations. We investigate the cellular and molecular mechanisms underlying such these specializations.
-
Zebrafish model for vascular biology: We use zebrafish model system, with extensive utilization of confocal microscopy, CRISPR-Cas9 mutagenesis, transgenesis and NGS analysis.
-
Vascular malformations and dysfunction models: Identification of the molecular correlates underlying vascular Malformation and dysfunction using the pre-clinical in vivo zebrafish model system.
-
Regeneration: Zebrafish is an excellent model for regeneration. We investigate revascularization of regenerating organs.
Key Publications
-
Das RN, Tevet Y, Safriel S, et al. Generation of specialized blood vessels via lymphatic transdifferentiation. Nature. 2022;606(7914):570-575. Highlighted article: Jeltsch M, Alitalo K. Lymphatic-to-blood vessel transdifferentiation in zebrafish. Nat Cardiovasc Res. 2022;1(6):539-541.
-
Das RN, Yaniv K. Discovering New Progenitor Cell Populations through Lineage Tracing and In Vivo Imaging. Cold Spring Harb Perspect Biol. 2020;12(10):a035618. Published 2020 Oct 1.
All publications link: https://vasculaturelab.wixsite.com/home/publications
Sanjeev Galande

We utilize various model systems to investigate the regulatory circuits involved in embryogenesis, regeneration, cancer, immunology, and neurogenesis. Our research primarily focuses on cellular signaling pathways and epigenetic modifiers that influence the chromatin landscape, ultimately shaping cellular phenotypes across different biological contexts.
Zebrafish Group
We are interested in exploring the molecular mechanisms that regulate cell fate decisions during different stages of zebrafish development. We focus on the genetic and epigenetic control of cell fate specification and cellular movements, particularly during gastrulation and organogenesis. By integrating genetic, imaging, and genome-wide approaches, we aim to identify and characterize novel vertebrate lineage-specific transcriptional regulators. Through investigating the interplay between chromatin modifications and transcriptional control, our research seeks to uncover the regulatory mechanisms shaping cell fate decisions.
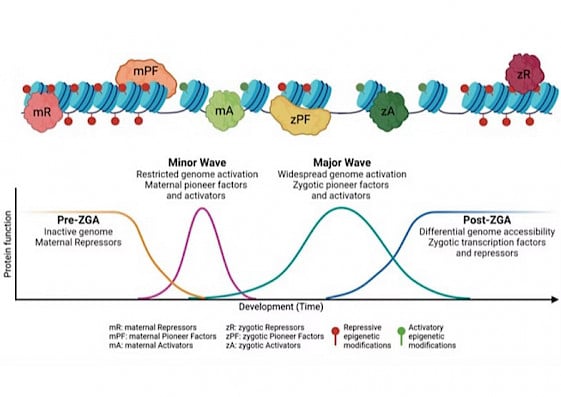 Precise gene expression is orchestrated by the interplay between multiple transcription factors and epigenomic marks, both of which can function as activators or repressors. These regulatory elements work in concert to modulate chromatin accessibility, leading to dynamic transitions between permissive and restrictive states. Such changes influence the recruitment of transcriptional machinery, ensuring proper gene regulation across different stages of development.
Precise gene expression is orchestrated by the interplay between multiple transcription factors and epigenomic marks, both of which can function as activators or repressors. These regulatory elements work in concert to modulate chromatin accessibility, leading to dynamic transitions between permissive and restrictive states. Such changes influence the recruitment of transcriptional machinery, ensuring proper gene regulation across different stages of development.Hydra Group
Hydra – the freshwater Cnidarian – is renowned for its robust regenerative potential. With a minimalistic body plan, well-defined regions of stem cell proliferation and differentiation, and established pharmacological and genetic perturbation methods, Hydra has a shining track record as a model to study the evolution of gene regulatory mechanisms and the consequences of their perturbation for functional comprehension. The Hydra research group is focused on unveiling conserved genetic and epigenetic regulatory mechanisms and pathways in morphogenesis and regeneration.
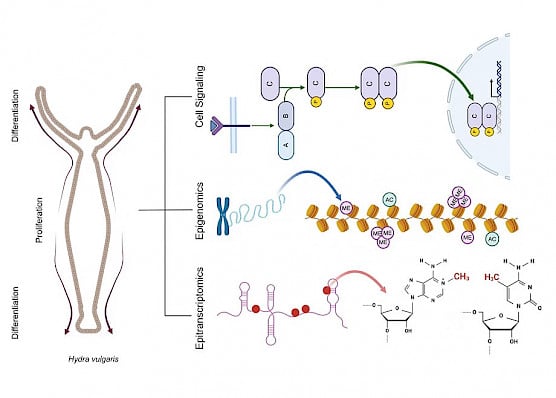
Projects/Highlights
-
Investigating the role of Satb2 as a gatekeeper of developmental transitions in early vertebrate embryogenesis.
-
Role of chromatin organization in controlling cell fate specification and morphogenesis during early vertebrate development.
-
Role of Sp1 in organogenesis, focusing on transcriptional control in the gastrointestinal tract.
-
Function of histone post-translational modifications in regulating gene expression during early embryogenesis, shaping developmental trajectories.
-
Wnt signalling cascade and downstream targets in head organizer formation
-
Enhancer switching in head organizer establishment
-
Role of tissue stiffness in Hydra regeneration and motility
-
Crosstalk of signalling pathways in tissue boundary maintenance
-
Evolutionary epitranscriptomics of cell fate determination
-
Epigenomic landscape determining promoter and enhancer dynamics
Key Publications
-
Karmodiya, K., Anamika, K., Muley, V., Pradhan, S.J., Bhide, Y. and Galande, S., 2014. Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development. Scientific Reports, 4:6076. doi: 10.1038/srep06076.
-
Pradhan, S.J., Reddy, P.C., Smutny, M., Sharma, A., Sako, K., Oak, M.S., Shah, R., Pal, M., Deshpande, O., Dsilva, G. and Tang, Y … & Galande, S., 2021. Satb2 acts as a gatekeeper for major developmental transitions during early vertebrate embryogenesis. Nature Communications, 12(1).:6094. doi: 10.1038/s41467-021-26234-7. PMID: 34667153.
-
Dsilva, G.J. and Galande, S., 2024. From sequence to consequence: Deciphering the complex cis-regulatory landscape. Journal of Biosciences, 49(2). DOI: 10.1007/s12038-024-00431-0.
-
Sharma, A., Dsilva, G.J., Deshpande, G. and Galande, S., 2024. Exploring the versatility of zygotic genome regulators: A comparative and functional analysis. Cell Reports, 43(9).
1-15 https://doi.org/10.1016/j.celrep.2024.114680.
All publications link: https://www.sglabepigenetics.com/publications
-
Gungi, A., Saha, S., Pal, M. and Galande, S., 2023. H4K20me1 plays a dual role in transcriptional regulation of regeneration and axis patterning in Hydra. Life Science Alliance, 6(5).
-
Unni, M.K., Reddy, P.C. and Galande, S., 2021. Identification of components of the Hippo pathway in Hydra and potential role of YAP in cell division and differentiation. Frontiers in Genetics, p.1903.
-
Pillai A, Gungi A, Reddy PC and Galande S (2021) Epigenetic Regulation in Hydra: Conserved and Divergent Roles. Front. Cell Dev. Biol. 9:663208. doi: 10.3389/fcell.2021.663208
-
Reddy, P. C., Gungi, A., Ubhe, S., & Galande, S. 2020. Epigenomic landscape of enhancer elements during Hydra head organizer formation. Epigenetics & Chromatin, 13(1), 43
-
Naik, S., Unni, M., Sinha, D., Rajput, S. S., Reddy, P. C., Kartvelishvily, E., Solomonov, I., Sagi, I., Chatterji, A., Patil, S., & Galande, S. 2020. Differential tissue stiffness of body column facilitates locomotion of Hydra on solid substrates. Journal of Experimental Biology, jeb.232702. Advance online publication.
-
Reddy, P.C., Gungi, A., Ubhe, S., Pradhan, S.J., Kolte, A. and Galande, S., 2019. Molecular signature of an ancient organizer regulated by Wnt/β-catenin signalling during primary body axis patterning in Hydra. Communications Biology, 2(1), pp.1-11.
Sumeet Pal Singh

Our lab explores the liver’s extraordinary regenerative capacity, uncovering the cellular and molecular mechanisms that enable recovery from injury, metabolic stress, and cancer. The liver can rebuild itself, but the efficiency of this process varies with age and disease. Using partial hepatectomy—a model that mimics a clinical procedure—we study how cell plasticity drives regeneration and have found this process is enhanced during growth spurts. We also investigate liver atrophy during prolonged starvation, mechanisms of liver failure in alcohol-induced damage, and strategies to enhance liver resilience in cancer.
By integrating single-cell transcriptomics, live imaging, and CRISPR-based gene editing, we map how liver cells adapt to injury and metabolic challenges. These insights provide a foundation for developing strategies to improve liver regeneration and resilience in disease contexts. Our work bridges developmental biology, metabolism, and regenerative medicine, offering new perspectives on organ repair and disease progression.
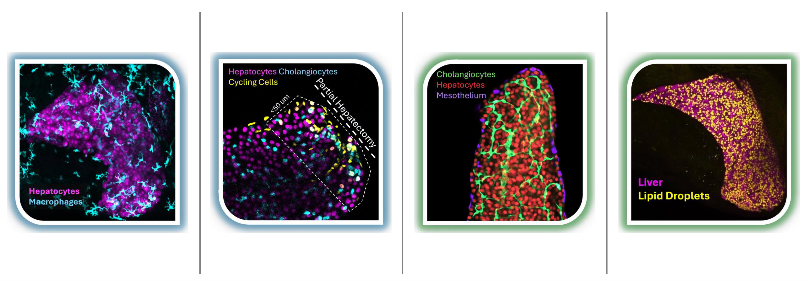
(From Left to Right) Inflammation in liver upon starvation; regenerative response to partial hepatectomy; hepatocytes and cholangiocytes in a growing liver; lipid droplets in the liver.
Projects/Highlights
- Cellular plasticity and regeneration in liver repair
- Starvation as a driver of liver atrophy and metabolic adaptation
- Mechanisms of liver failure and resilience in cancer and alcohol-induced damage
- Single-cell and imaging approaches to dissect liver regeneration
- Intersection of developmental biology, metabolism, and regenerative medicine
Key Publications
- Cholangiocytes contribute to hepatocyte regeneration after partial liver injury during growth spurt in zebrafish.
bioRxiv (https://www.biorxiv.org/content/10.1101/2025.01.09.629100v1)
Sema Elif Eski, Jiarui Mi, Macarena Pozo-Morales, Gabriel Garnik Hovhannisyan, Camille Perazzolo, Rita Manco, Imane Ez-Zammoury, Dev Barbhaya, Anne Lefort, Frederick Libert, Federico Marini, Esteban N. Gurzov, Olov Andersson, Sumeet Pal Singh - Starvation resistant cavefish reveal conserved mechanisms of starvation-induced hepatic lipotoxicity Life Science Alliance (https://www.life-science-alliance.org/content/7/5/e202302458) Macarena Pozo-Morales*, Ansa E Cobham*, Cielo Centola, Mary Cathleen McKinney, Peiduo Liu, Camille Perazzolo, Anne Lefort, Frederick Libert, Hua Bai, Nicolas Rohner§, Sumeet Pal Singh§
* Co-Authors § Co-Corresponding author.
- In vivo imaging of calcium dynamics in zebrafish hepatocytes.Hepatology (https://journals.lww.com/hep/Abstract/2023/03000/In_vivo_imaging_of_calcium_dynamics_in_zebrafish.10.aspx) Macarena Pozo-Morales, Inés Garteizgogeascoa, Camille Perazzolo, Juhoon So, Donghun Shin, Sumeet Pal Singh.
- A single-cell atlas of de novo β-cell regeneration reveals the contribution of hybrid β/δ cells to diabetes recovery in zebrafish.Development (https://doi.org/10.1242/dev.199853) Sumeet Pal Singh*, Prateek Chawla*, Alisa Hnatiuk, Margrit Kamel, Luis Delgadillo Silva, Bastiaan Spanjard, Sema Elif Eski, Sharan Janjuha, Pedro Olivares, Oezge Kayisoglu, Fabian Rost, Juliane Bläsche, Annekathrin Kränkel, Andreas Petzold, Thomas Kurth, Susanne Reinhardt, Jan Philipp Junker, Nikolay Ninov
* Co-Authors
- Single-cell transcriptome analysis reveals thyrocyte diversity in the zebrafish thyroid gland EMBO reports (https://doi.org/10.15252/embr.202050612)
Pierre Gillotay, Meghna P Shankar, Sema Elif Eski, Macarena Pozo-Morales, Inés Garteizgogeascoa, Susanne Reinhardt, Annekathrin Kraenkel, Juliane Blaesche, Andreas Petzold, Nikolay Ninov, Gokul Kesavan, Christian Lange, Michael Brand, Vincent Detours, Sabine Costagliola§, Sumeet Pal Singh§
§ Co-Corresponding author
All publications link: https://sumeetpalsingh.github.io/publications